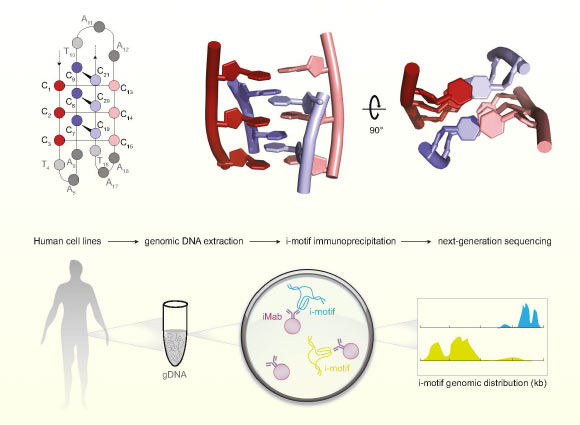
The so-called i-motif is a knot-like DNA structure that forms in the nuclei of human cells and is thought to provide important genome control. Garvan Institute of Medical Research Other studies have used immunoprecipitation and next-generation sequencing to identify i-motif structures in human DNA.
Peña Martinez othersIn total, we observed 53,000 i-motifs across three human cell lines (MCF7, U2OS, and HEK293T). Image courtesy of Peña Martínez. others., doi: 10.1038/s44318-024-00210-5.
The I motif is a DNA structure that differs from the iconic double helix shape.
These form when runs of cytosine letters on the same DNA strand pair up with each other to form a four-stranded twisted structure that juts out from the double helix.
In 2018, scientists at the Garvan Institute of Medical Research were the first to successfully directly visualize i-motifs inside living human cells, using new antibody tools they developed to recognise and bind to the i-motifs.
The new study expands on these findings by using the antibody to identify the location of i-motifs throughout the genome.
“In this study, we have mapped more than 50,000 i-motif sites in the human genome that are found in all three cell types we looked at,” said Professor Daniel Crist from the Garvan Institute of Medical Research, lead author of the study.
“This is a surprisingly high number for a DNA structure whose presence in cells was once a matter of debate.”
“Our findings confirm that the i-motif is not just an object of laboratory study, but is widespread and likely plays an important role in genome function.”
The researchers found that i-motifs are not scattered randomly, but are concentrated in important functional regions of the genome, including those that control gene activity.
“We found that the i-motif is associated with genes that are highly active at specific times in the cell cycle,” said lead author Cristian David Peña Martinez, PhD, also of the Garvan Medical Institute.
“This suggests that it plays a dynamic role in regulating gene activity.”
“We also discovered that i-motifs are formed in the promoter regions of cancer genes. For example, MYC Oncogenes encode one of cancer’s most notoriously ‘untreatable’ targets.”
“This opens up exciting opportunities to target disease-related genes through i-motif structures.”
“The widespread presence of the i-motif near these 'holy grail' sequences implicated in hard-to-treat cancers opens up new possibilities for novel diagnostic and therapeutic approaches,” said study co-author Sarah Kummerfeld, PhD, a researcher at the Garvan Medical Institute.
“It may be possible to design drugs that target the i-motif to affect gene expression, potentially expanding current treatment options.”
Team result Published in EMBO Journal.
_____
Christian David Peña Martinez othersi-motif structures are widely distributed in human genomic DNA. Embo JPublished online August 29, 2024, doi: 10.1038/s44318-024-00210-5
Source: www.sci.news