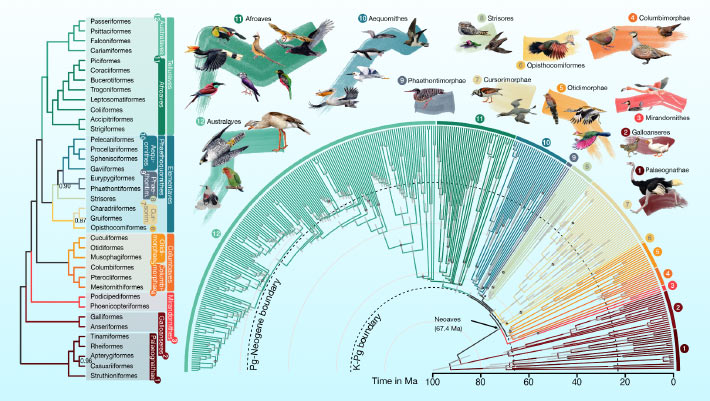
The latest genealogy is detailed in two supplementary papers published today. journal Nature And that Proceedings of the National Academy of Sciences, researchers have uncovered patterns in the evolutionary history of birds after the massive mass extinction event that wiped out the dinosaurs 66 million years ago. The authors observed rapid increases in effective population size, replacement rate, and relative brain size in early birds, and found that new adaptive mechanisms that drove bird diversification in the aftermath of this pivotal event. Shined a light. The researchers also took a closer look at one branch of the new family tree and found that flamingos and pigeons are more distantly related than previous genome-wide analyzes had shown.
The latest bird family tree outlining 93 million years of evolutionary relationships among 363 bird species. Image credit: Jon Fjeldså / Josefin Stiller.
“Our goal is to reconstruct the entire evolutionary history of all birds,” said Professor Siavash Milarab, a researcher at the University of California, San Diego.
This work is part of that Bird 10,000 Genomes (B10K) ProjectThis is a multi-institutional effort led by the University of Copenhagen, Zhejiang University, and the University of California, San Diego, with the aim of producing draft genome sequences for approximately 10,500 extant bird species.
At the heart of these studies is a suite of algorithms known as ASTRAL, developed by Professor Miralove and colleagues to infer evolutionary relationships with unprecedented scalability, accuracy, and speed.
By harnessing the power of these algorithms, we integrated genomic data from over 60,000 genomic regions and provided a robust statistical foundation for our analysis.
The researchers then examined the evolutionary history of individual segments across the genome.
From there, they pieced together a mosaic of gene trees and compiled them into a comprehensive species tree.
This meticulous approach has allowed researchers to construct new and improved bird genealogies that depict complex divergence events with remarkable accuracy and detail, even in the face of historical uncertainty. I did.
“We found that our method, which adds tens of thousands of genes to the analysis, is indeed necessary to unravel the evolutionary relationships between bird species,” Professor Miralove said.
“We really need all the genomic data to reconstruct with a high degree of confidence what happened during this period of time, 65 to 67 million years ago.”
The scientists also looked at the impact of different genome sampling methods on the accuracy of the tree.
They showed that to reconstruct this evolutionary history, it is important to combine two strategies: sequence many genes in each species and sequence many species. Ta.
“Because we used both strategies in combination, we were able to test which approach has a stronger impact on phylogenetic reconstructions,” said Professor Josephine Stiller from the University of Copenhagen.
We found that it is more important to sample many gene sequences from each organism than to sample from a wider range of species, but the latter method does not allow us to determine when different groups evolved. It was helpful to know. ”

mira love other. They took a closer look at one branch of the updated bird family tree and found that groups including flamingos and pigeons are more distantly related than previous genome-wide analyzes had shown. We attributed the results to an abnormal region on chromosome 4.Image credits: Ed Braun / Daniel J. Field / Siavash Miarab
With the help of advanced computational techniques, the researchers were also able to shed light on anomalies discovered in previous studies. The theory is that a particular part of a chromosome in the bird's genome remained unchanged and blank for millions of years. Description of expected genetic recombination patterns.
“Ten years ago, we put together a family tree. Neo Avesthe group that includes the vast majority of bird species,” said Professor Edward Brown of the University of Florida.
“Based on the genomes of 48 species, we divided neoabees into two broad categories: pigeons and flamingos in one group, and all the rest in the other.”
“This year, when we repeated the same analysis with 363 species, a different family tree emerged that divided pigeons and flamingos into two distinct groups.”
“Given two mutually exclusive family trees, I looked for an explanation that would allow me to determine which family tree was correct.”
“When we looked at individual genes and which trees they supported, it suddenly dawned on us that all the genes that support old trees were all in one place. That's how it all started. “It was,” he explained.
“When we investigated this site, we realized that it was a place where sexual reproduction had been occurring for millions of years, but it wasn't as mixed.”
“Just like humans, birds combine the genes of their father and mother to create the next generation.”
“But in birds and humans alike, when creating sperm and eggs, we first mix together genes inherited from both parents.”
“This process, called recombination, maximizes the genetic diversity of a species by ensuring that no two siblings are exactly alike.”
The authors found evidence that parts of bird chromosomes suppressed this recombination process for millions of years after the dinosaurs went extinct.
It is unclear whether extinction events and genomic abnormalities are related.
They found that flamingos and pigeons resemble each other in this frozen chunk of DNA.
However, when the complete genomes were considered, it became clear that the two groups were more distantly related.
“What is surprising is that this period of recombination suppression can mislead the analysis,” says Professor Brown.
“And because that can mislead the analysis, it was actually detectable more than 60 million years in the future. That's the cool thing about it.”
“Such mysteries may also be hidden in the genomes of other organisms.”
“We discovered this misleading region of birds because we put a lot of energy into deciphering their genomes.”
“I think there are similar cases in other species that are unknown at this time.”
_____
J. Stiller other. 2024. The complexity of bird evolution revealed by family-level genomes. Naturein press.
Siavash Milarab other. 2024. Suppressed recombinant regions mislead neoavian phylogenomics. PNAS 121: e2319506121; doi: 10.1073/pnas.2319506121
Source: www.sci.news